Genes act in concert to perform various functions in cells and tissues. Understanding the function of genes at the module (or set) level is a critical step in our understanding of bimolecular processes. The growing body of functional genomics data enables the systematic screening and collection of results oriented around a set of interest. BEAST enables a researcher to easily harvest such information and perform various concept enrichment analyses and transformations.
The UCSC Biomolecular Entities and Sets Tools (BEAST) is an interactive web application that allows biologists to search, define and filter sets of molecular entities such as genes, proteins, small molecule drugs, and mRNAs. These sets can then be further manipulated and analyzed using the BEAST tool. Users may import their own sets of biological entities and seamlessly compare them any existing sets in the BEAST native database.
The BEAST supports set overlaps, transformations, and statistical tests for data mining.
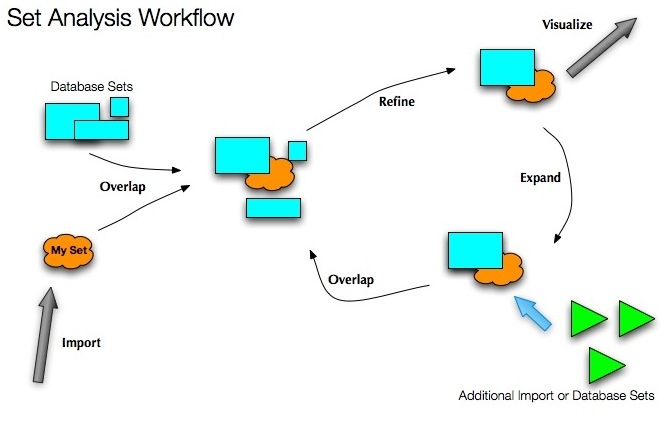
Set analysis workflow:

BEAST users can browse the entire Gene Ontology database for multiple organisms, either by viewing an interactive hierarchical tree, or by searching on the GO descriptions themselves.
Selected subsets of GO are then imported into the users workspace, where all sets and their members can be viewed via a set-membership heatmap.
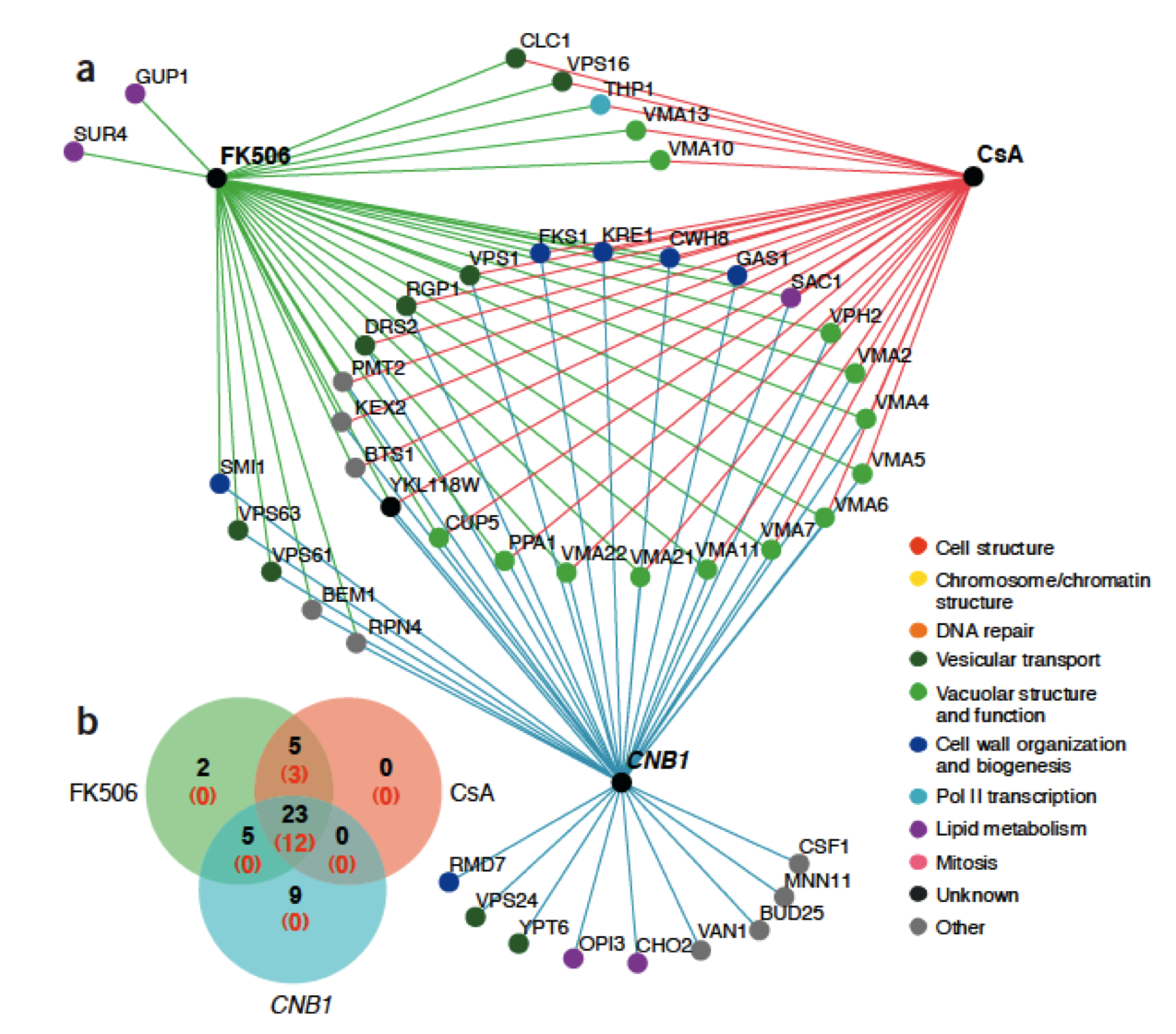
Genes act together to produce a phenotype for the cell. By studying the survivability of many combinations of double-mutants in yeast, the Boone lab has managed to construct a interaction map of many of the non-vital genes.
Each hub gene can be thought of as a set, with its members the genes who, when removed along with that hub gene, produce a less survivable phenotype. These sets are referred to as the synthetic lethal neighborhoods (SLNs) of a given hub gene.
Users who are studying the effects of a chemical on specific genetic pathways may want to test the effects of that chemical on a set of yeast knockouts. This will generate data consisting of a set of yeast genes that are sensitive to that compound, which can then be uploaded to the BEAST and compared with the Boone SLNs. The overlaps between the chemcial and SLN sets can be visualized as a heatmap in the BEAST, allowing users to quickly target hub genes that may interact with their chemcial compound.
As a follow up to their target predictions, the users may then overlap the SLNs of the predicted target genes with a subcategory of GeneOntology, to gain insight into the target pathways or mechanisms of action of the target drugs, or compare their interactions with known drug targets from DrugBank, or other databases pre-loaded into the BEAST application.
Step 1: Add sets to your workspace
Go the the BEAST Website and click on the plus sign next to the 'Hillenmeyer Chemical - Knockout
Interaction Sets' line.
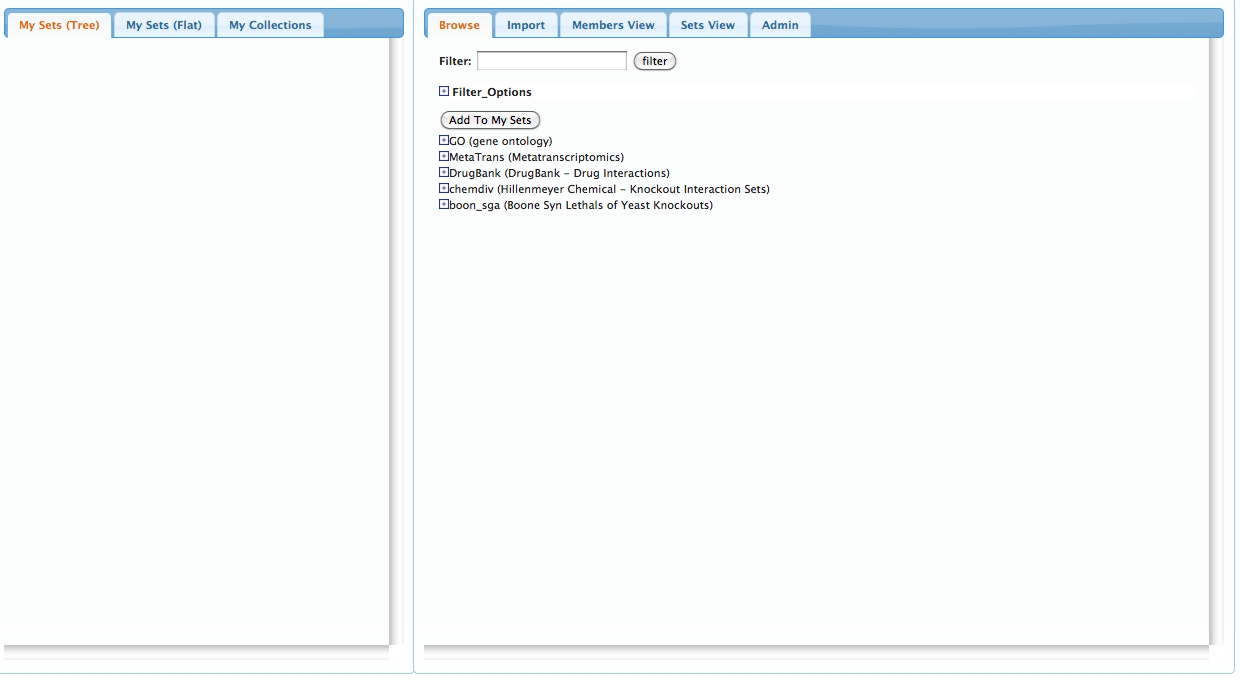
You should now see a drop-down list of chemical compounds, each representing the sensitivity set of genes for that compound.
Scroll down and try selecting 'rapamycin' at several doses, as well as the drug 'nocozodole' for comparison, as well
as a few of the 'minimal-media' checkboxes to use as a control. Scroll back to the top and hit the 'Add To My Sets'
button: you should see the left-hand pannel display the sets you just added.
Now, go back to the top of the right-hand pannel, hide the 'chemdiv' drop-down box by clicking on the '-' sign next to it, and select the 'Boone Syn Lethals of Yeast Knockouts' drop-down menu. You'll see a number of Yeast 'query' genes appear, which represent the synthetic lethal set of each, from the Boone map. Select a handful of genes, and make sure to include PEP12 and COG8 among your choices--now hit the 'Add To My Sets' button at the top again to import those choices into the left-hand pannel. Your UI should now look something like this:
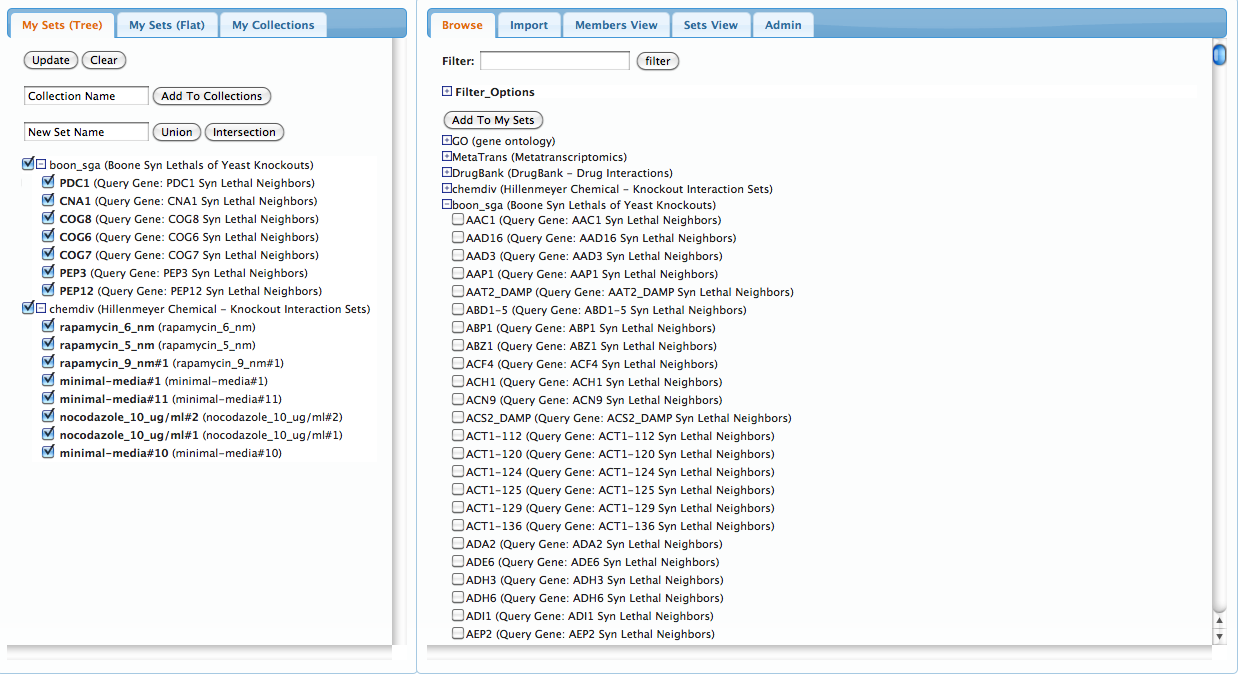
Step 2: Create collections Collections are sets of sets with a common theme or purpose. In this tutorial, we would like to compare the synthetic lethal neighborhoods of our query genes (from the Boone map) with chemical sensitivity signatures from the Hillenmeyer dataset.
On the left pannel, select just the synthetic lethal genes by selecting the top-level 'Boone Syn...' checkbox and unchecking the 'chemdiv' checkbox. Type a name for this new collection into the first text area and click the 'Add To Collections' button:
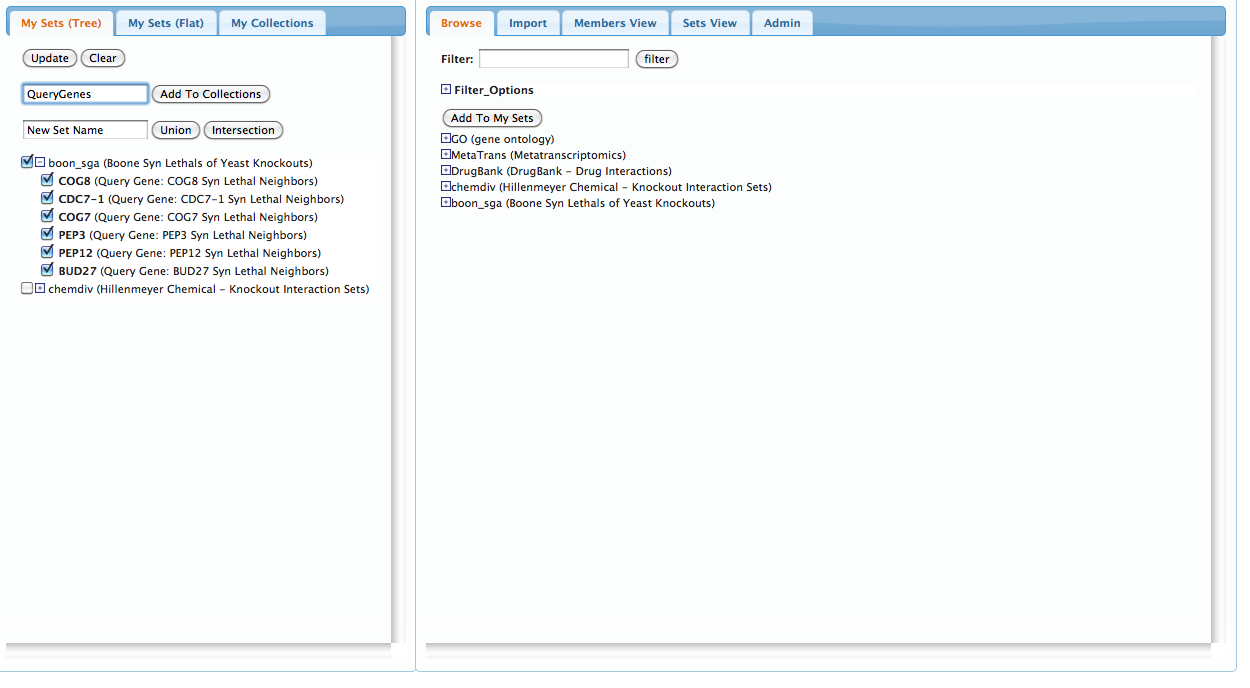
Now, do the same for the 'chemdiv' sets to create a collection of chemical compounds:
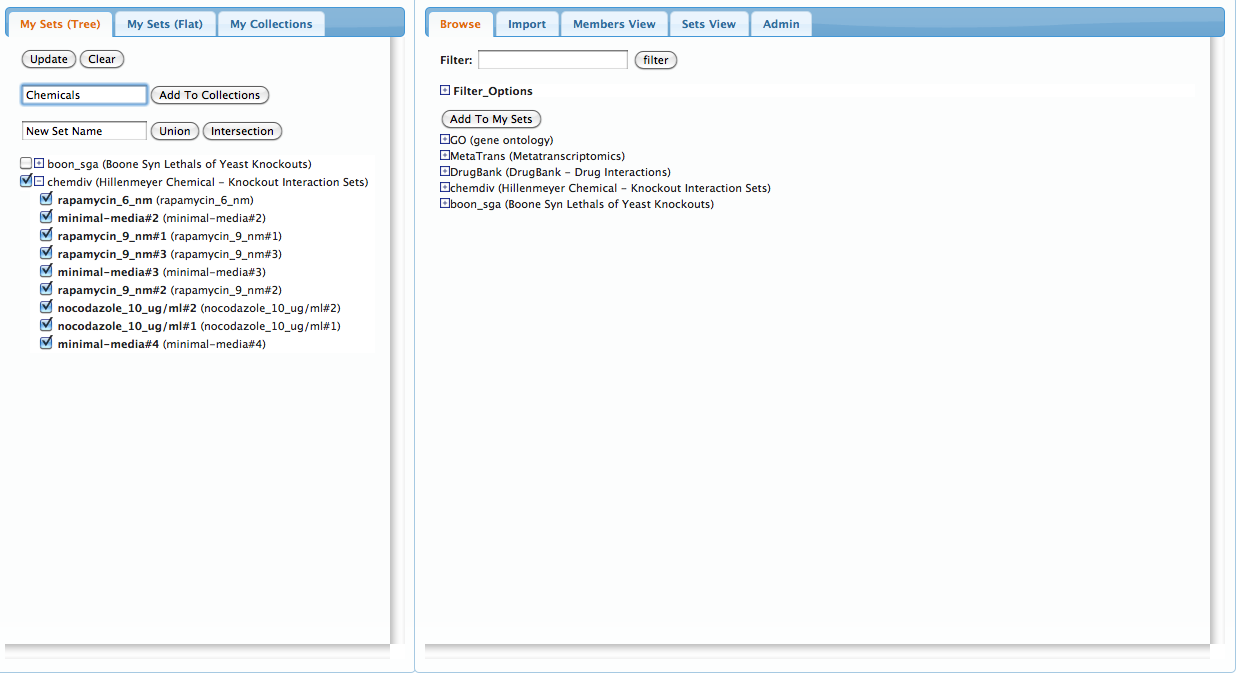
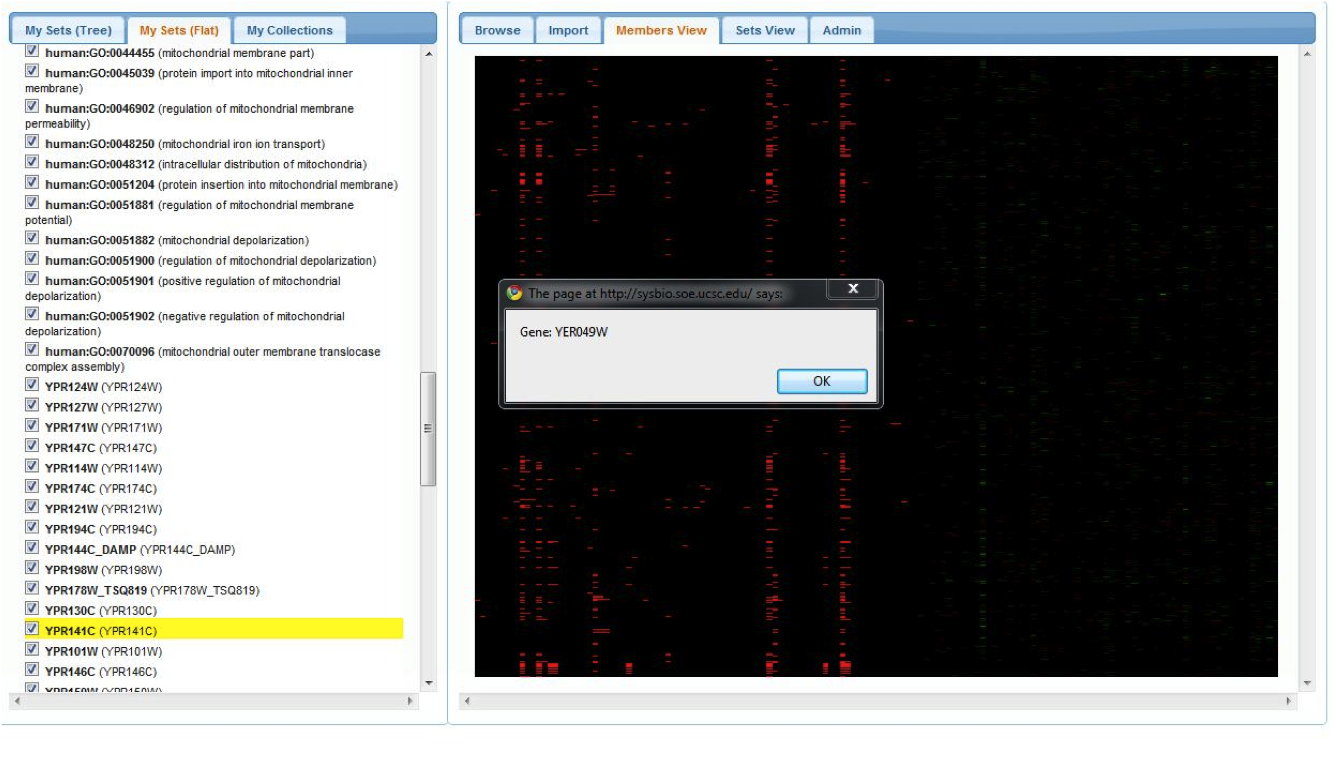
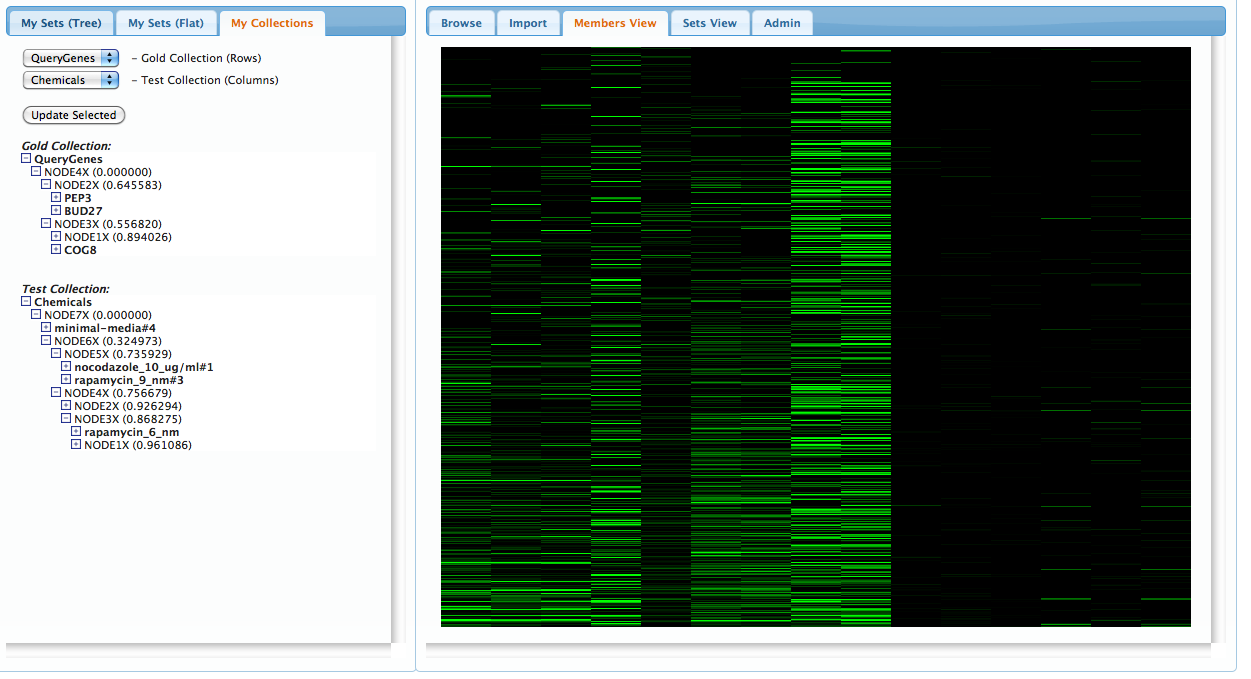
Before continuing, we can view all of the sets in our current workspace by selecting the 'Members View' pannel on the right. The rows represent gene-memberships for the set of all genes in the union of all our current sets, while columns represent each set. Clicking on a column will highlight the corresponding set on the left pannel:
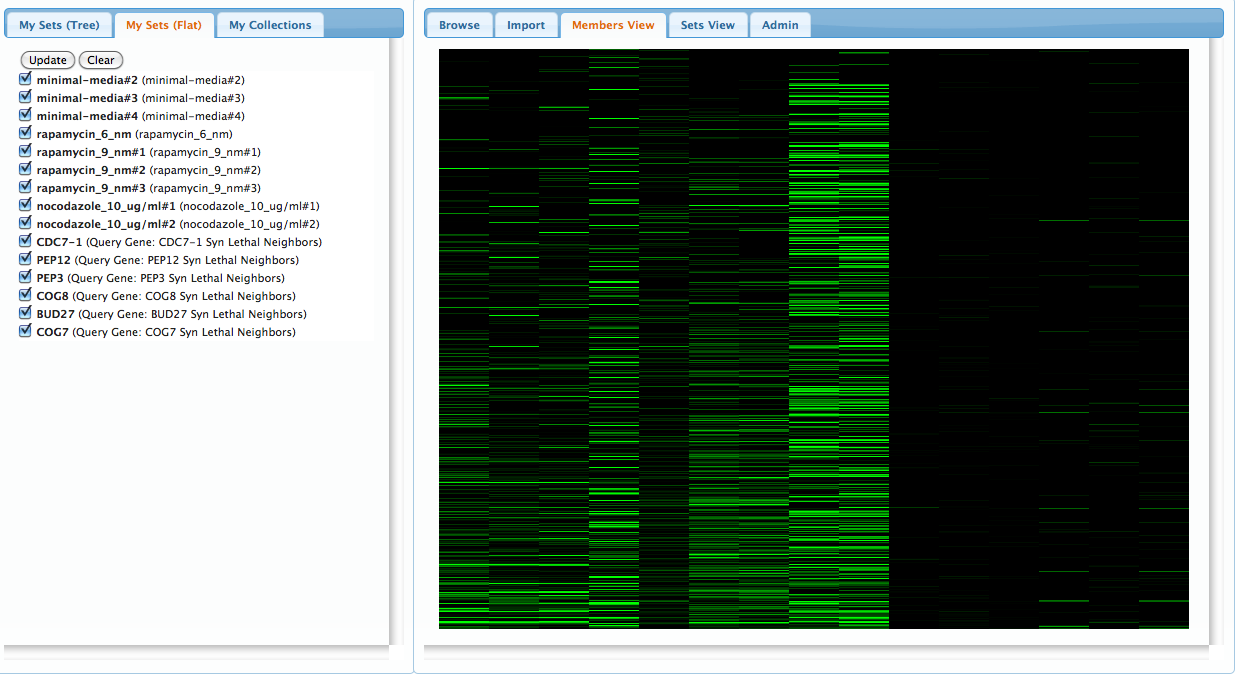
Step 3: compare collections
We wish to compare the two collections we created in the previous step to see if any of the sets in the Boone query gene collection are similar to chemical signatures in the Hillenmeyer chemical-sensitivity set in the second collection. This will tell us if any of the candidate chemical compounds are likely to target these specific genes.Click on the 'My Collections' pannel on the left and select the 'QueryGenes' collection in the first drop-down box. Choose the 'Chemicals' collection for the second drop-down box, and then click the 'Update Selected' button. The server will perform a heirarchical clustering operation on each collection and display the results in the area below:
Now, on the right hand pannel, click the 'Sets View' tab to bring up a heatmap comparison of the two collections. You'll see a grid with rows representing each query gene set, and columns representing each compound sensitivity set. The shade in each box represents the strength of the overlap for each pair of sets: the redder the color, the stronger the overlap according to the hypergeometric (Fisher's exact) test. Note, in the example below, the strong overlap between the right-most column and the fourth row down: click on this square and the corresponding chemical and gene will be highlighted in the left-hand pannel:
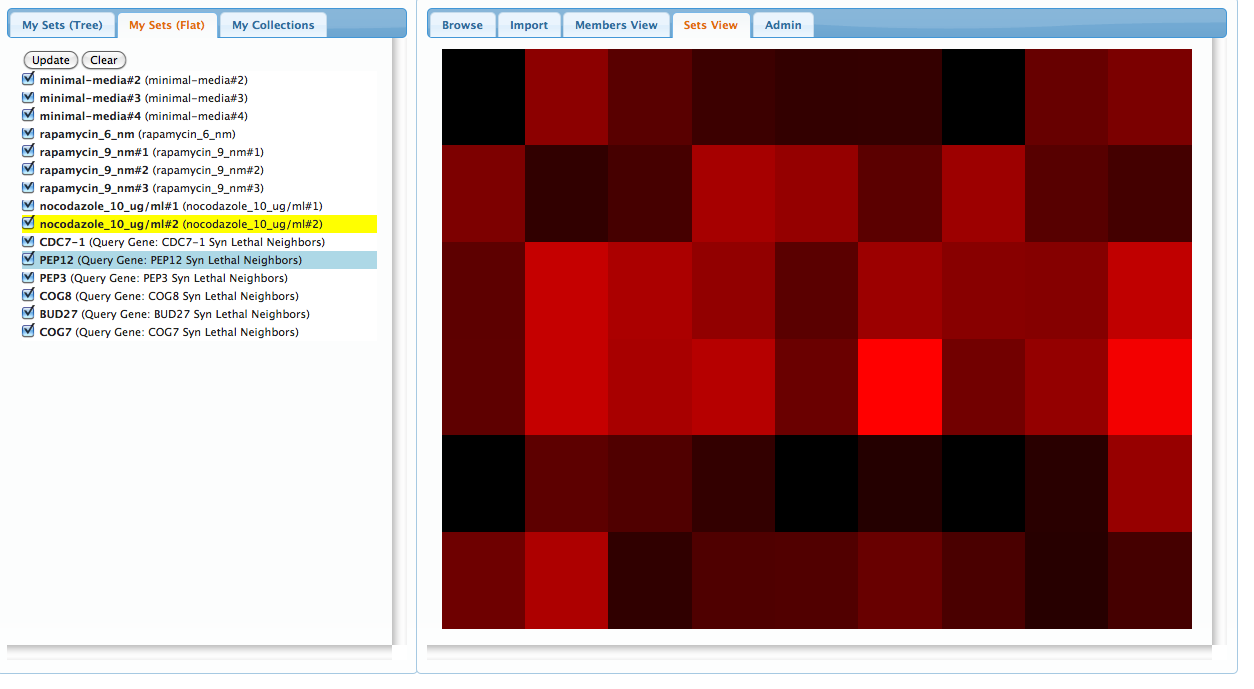
This is a useful result: we've found evidence that nocodazole targets the PEP12 gene. However, looking at the fourth row, we can also see that at least one of the other compounds has a strong overlap with the PEP12 gene (4th row), so it's possibly that PEP12 is sensitive to many compounds, which reduces the specificity of our result. At this point, the user would be wise to refine their test by bringing in other query genes and chemical sensitivity for a more detailed comparison. Testing these sets against Gene-Ontology categories may be useful as well, or you can create your own sets and upload them through the 'Import' tab
Future directions include: - The addition of several popular biological entity databases, including KEGG and Reactome.
- Multiple modes of determining set-overlap significance will be available, including various statistical measures as well as simple set coverage metrics.
- Additional visualization options will be available, including possible zoom features, and export to Java TreeView.
Customizable sessions and the ability to save your search and analysis results in our database for quick retrieval.
Integration with the UCSC Interaction Browser, the Stem Cell Browser and other set-related tools.